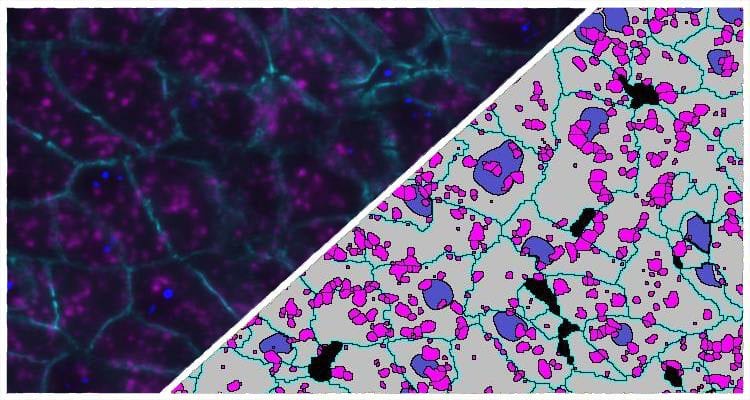
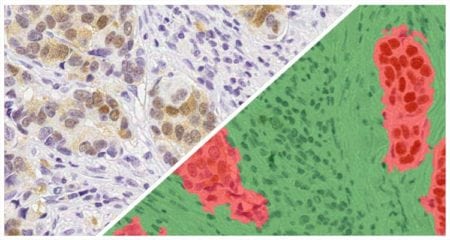
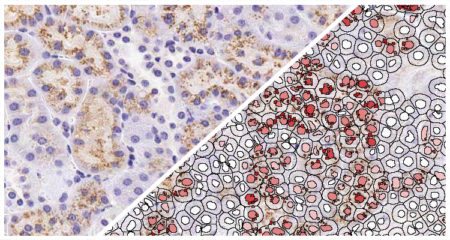
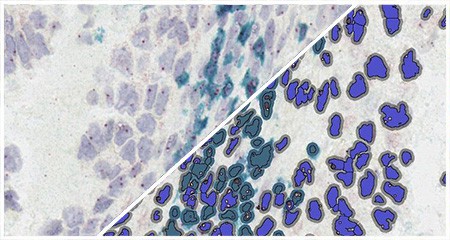
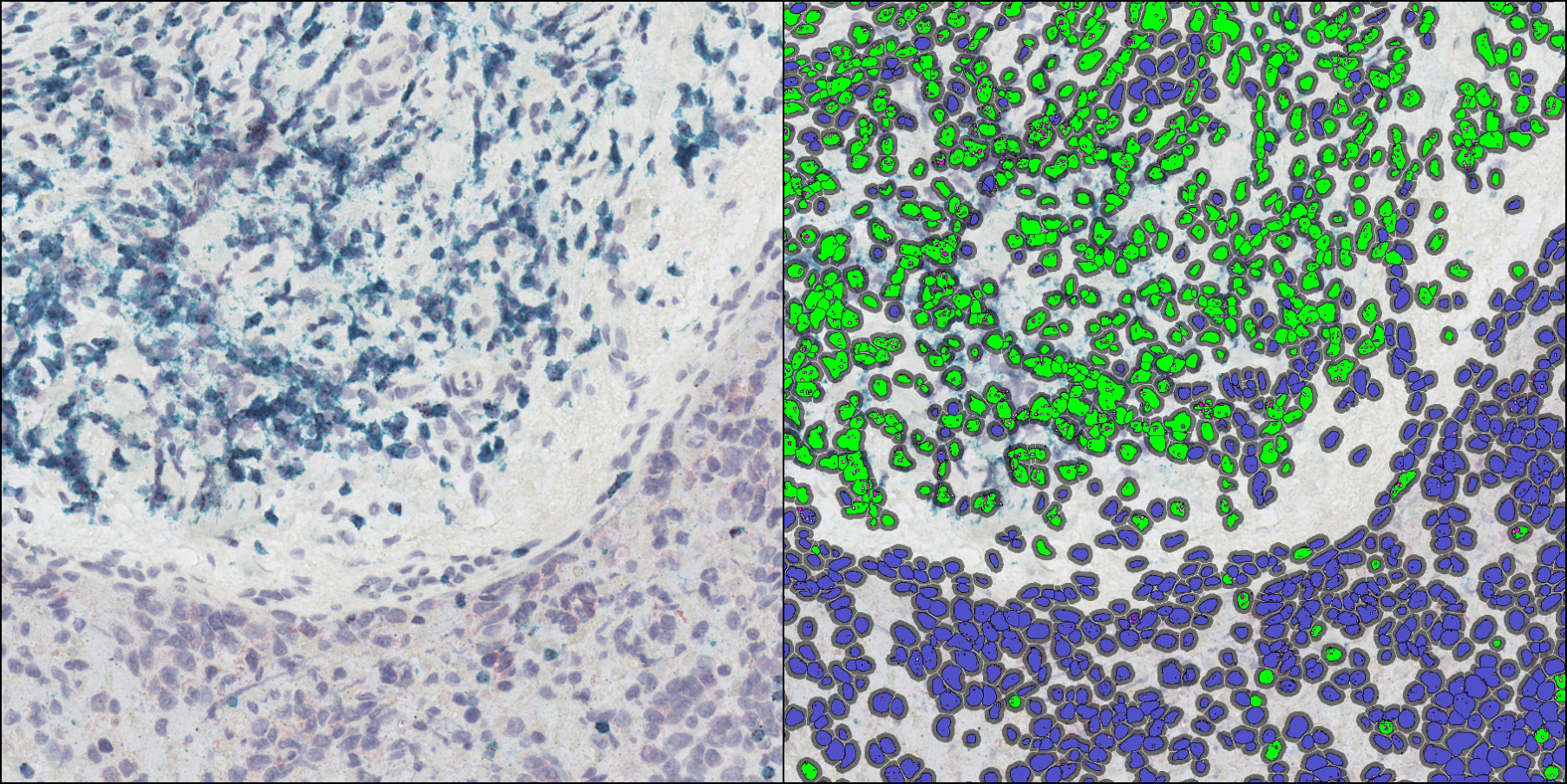
Indica Labs FISH-IF quantification module measures any number of fluorescently-labeled DNA/RNA ISH probes and immunofluorescent (IF) protein biomarkers on a cell-by-cell basis. This allows the user to rapidly contextualize the corresponding protein and gene expression profile of every cell across the tissue. HALO® FISH-IF analysis is designed to work with an unlimited number of fluorescent protein biomarkers and probes.
File formats supported by the HALO image analysis platform:
- Non-proprietary (JPG, TIF, OME.TIFF)
- Nikon (ND2)
- 3D Histech (MRXS)
- Akoya (QPTIFF, component TIFF)
- Olympus / Evident (VSI)
- Hamamatsu (NDPI, NDPIS)
- Aperio (SVS, AFI)
- Zeiss (CZI)
- Leica (SCN, LIF)
- Ventana (BIF)
- Philips (iSyntax, i2Syntax)
- KFBIO (KFB, KFBF)
- DICOM (DCM*)
*whole-slide images
Quantitative Image Analysis of a Combined FISH-IF Assay
Learn how to detect protein and RNA in a single assay using ACD’s Co-Detection workflow and HALO image analysis.
Submit the form below to view the requested document

RNAscope® Publications with the HALO® Image Analysis Platform
Read this publications list to see examples of research in areas from neuroscience to oncology using HALO to analyze RNAscope images.
Submit the form below to view the requested document

Quantitative RNAscope Image Analysis Guide
From experimental design considerations to optimized setup of HALO image analysis parameters, our guide will help take your quantitative RNAscope™ image analysis to the next level.
Submit the form below to view the requested document

Getting Started with RNAscope™ Image Analysis in HALO®
28 March 2023 | Join us for this 1-hour webinar to see a live demonstration of RNAscope image analysis using the HALO® platform from Indica
Masterclass Webinar: Optimizing RNAscope Image Analysis
05 May 2022 | In this 60-min webinar, Dr. Ghislaine Lioux will present solutions to common RNAscope image analysis challenges including how to optimize

Whole-slide Quantitative RNAscope Image Analysis: Applications and Methods
20 April 2022 | In this 2-hour event, learn from three experts in RNAscope image analysis with HALO® as they present their scientific research, share

HALO Image Analysis Masterclass Series: February-March 2021
Spring of 2021 | Indica Labs is excited to continue our HALO® Masterclass Webinar Series this winter. Each masterclass webinar will offer a deep dive
Publication Spotlight
The table below includes publications that cite the FISH-IF and ISH-IHC modules.
Your publication not on the list? Drop us an email to let us know about it!
| Title | Authors | Year | Journal | Application | HALO Modules | Product |
|---|---|---|---|---|---|---|
| Detection of EpsteinñBarr Virus in Periodontitis: A Review of Methodological Approaches | Tonoyan L, Chevalier M, Vincent-Bungnas S, Marsault R, Doglio A | 2021 | Mircroorganisms | Review | ISH/FISH, ISH-IHC/FISH-IF | HALO |
| Detection of engineered T cells in FFPE tissue by multiplex in situ hybridization and immunohistochemistry | Wright JH, Huang L-Y, Weaver S, Archila LD, McAfee MS, Hirayama AV, Chapuis AG, Bleakley M, Rongvaux A, Turtle CJ, Chanthaphavong S, Campbell JS, Pierce RH | 2020 | Journal of Immunological Methods | Immunology | ISH/FISH, ISH-IHC/FISH-IF | HALO |
| Epicardium-derived cells organize through tight junctions to replenish cardiac muscle in salamanders | Eroglu E, Yen C, Witman N, Elewa A, Araus A, Wang H, Szattler T, Umeano C, Sohlmer J, Goedel A, Simon A, Chien K | 2022 | Nature Cell Biology | Myology | ISH/FISH, ISH-IHC/FISH-IF | HALO |
| Satellite repeat RNA expression in epithelial ovarian cancer associates with a tumor immunosuppressive phenotype | Porter R, Sun S, Flores M, Berzolla E, You E, Phillips I, KC N, Desai N, Tai E, Szabolcs A, Lang E, Pankaj A, Raabe M, Thapar V, Xu K, Nieman L, Rabe D, Kolin D, Stover E, Pepin D, Stott S, Deshpande V, Liu J, Solovyov A, Matulonis U, Greenbaum B, Ting D | 2022 | The Journal of Clinical Investigation | Immuno-oncology | ISH/FISH, ISH-IHC/FISH-IF | HALO |
| Systemic Nos2 Depletion and Cox inhibition limits TNBC disease progression and alters lymphoid cell spatial orientation and density | Somasundaram V, Ridnour L, Cheng R, Walke A, Kedei N, Bhattacharyya D, Wink A, Edmondson E, Butcher D, Warner A, Dorsey T, Scheiblin D, Heinz W, Bryant R, Kinders R, Lipkowitz S, Wong S, Pore M, Hewitt S, McVicar D, Anderson S, Chang J, Glynn S, Ambs S, Lockett S, Wink D | 2022 | Redox Biology | Oncology | Classifer, ISH/FISH, Spatial Analysis, Registration, ISH-IHC/FISH-IF | HALO |
| Annexin A2/TLR2/MYD88 pathway induces arginase 1 expression in tumor-associated neutrophils | Zhang H, Zhu X, Friesen T, Kwak J, Pisarenko T, Mekvanich S, Velasco M, Randolph T, Kargl J, Houghton A | 2022 | Journal of Clinical Investigation | Immuno-oncology | Spatial Analysis, ISH-IHC/FISH-IF | HALO |
| Spatiotemporal transcriptome analysis reveals critical roles for mechano-sensing genes at the border zone in remodeling after myocardial infarction | Yamada S, Ko T, Hatsuse S, Nomura S, Zhang B, Dai Z, Inoue S, Kubota M, Sawami K, Yamada T, Sassa T, Katagiri M, Fujita K, Katoh M, Ito M, Harada M, Toko H, Takeda N, Morita H, Aburatani H, Komuro I | 2022 | Nature Cardiovascular Research | Myology | ISH-IHC/FISH-IF | HALO |
| Loss of functional System x-c uncouples aberrant postnatal neurogenesis from epileptogenesis in the hippocampus of Kcna1-KO mice | Aloi M, Thompson S, Quartapella N, Noebels J | 2022 | Cell Reports | Neuroscience | ISH-IHC/FISH-IF | HALO |
| Multiparameter immunohistochemistry analysis of HIV DNA, RNA and immune checkpoints in lymph node tissue | Richardson Z, Deleage C, Tutuka C, Walkiewica M, Del Rio-Estrada P, Pascoe R, Evans V, Reyesteran G, Gonzales M, Roberts-Thomson S, Gonzalez-Navarro M, Torres-Ruiz, F, Estes J, Lewin S, Cameron P | 2021 | Journal of Immunological Methods | Infectious Disease | ISH-IHC/FISH-IF | HALO |
| The potassium channel auxiliary subunit Kv_2 (Kcnab2) regulates Kv1 channels and dopamine neuron firing | Yee J, Rastani A, Soden M | 2022 | Journal of Neurophysiology | Neuroscience | ISH-IHC/FISH-IF | HALO |
| Transcriptional vulnerabilities of striatal neurons in human and rodent models of Huntington’s disease | Matsushima A, Pineda S, Crittenden J, Lee H, Galani K, Mantero J, Tombaugh G, Kellis M, Heiman M, Graybiel A | 2023 | Nature Communications | Neuroscience | Area Quantification, ISH-IHC/FISH-IF | HALO |
| Exploration of spatial heterogeneity of tumor microenvironment in nasopharyngeal carcinoma via transcriptional digital spatial profiling | Wang L, Wang D, Zeng X, Zhang Q, Wu H, Liu J, Wang Y, Liu G, Poan Y | 2023 | International Journal of Biological Sciences | Oncology | Area Quantification, ISH-IHC/FISH-IF | HALO |
Related HALO Modules
Separate multiple tissue classes across a tissue using a learn-by-example approach. Can be used in conjunction with all other modules (fluorescent and brightfield) to select specific tissue classes for further analysis.
Learn MoreQuantify spot number, optical density and co-expression of one or two brightfield RNA or DNA probes on a per cell basis. Includes support for brightfield RNAscope assays.
Learn MoreQuantify IHC positivity and spot number, optical density and co-expression of one or two brightfield RNA or DNA probes on a per cell basis. Includes support for brightfield RNAscope assays.
Learn MoreUse the arrows above to view additional related modules
Want to Learn More?
Fill out the form below to request information about any of our software products.
You can also drop us an email at info@indicalab.com