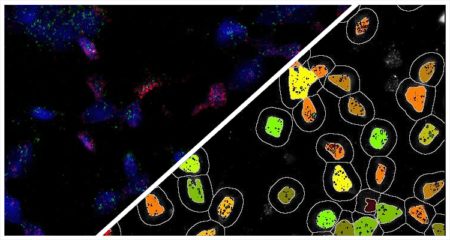
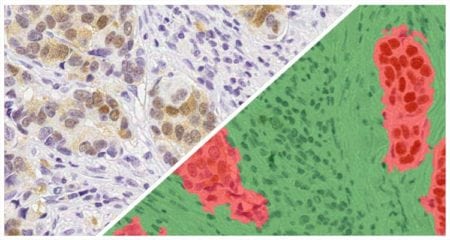
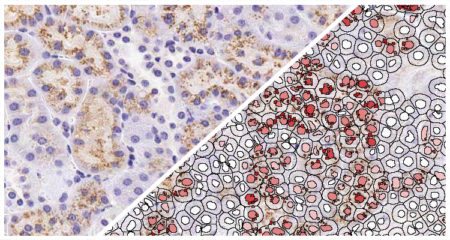
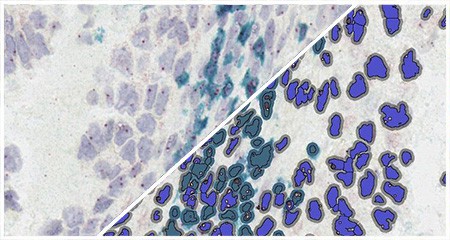
Indica Labs’ FISH module quantifies an unlimited number of DNA or RNA FISH probes on a cell-by-cell basis. This analysis module can be used with most multiplex RNA FISH assays including RNAscope® and supports the H-score protocol for RNAscope® recommended by the manufacturer, ACD, a Bio-techne brand.
File formats supported by the HALO image analysis platform:
- Non-proprietary (JPG, TIF, OME.TIFF)
- Nikon (ND2)
- 3D Histech (MRXS)
- Akoya (QPTIFF, component TIFF)
- Olympus / Evident (VSI)
- Hamamatsu (NDPI, NDPIS)
- Aperio (SVS, AFI)
- Zeiss (CZI)
- Leica (SCN, LIF)
- Ventana (BIF)
- Philips (iSyntax, i2Syntax)
- KFBIO (KFB, KFBF)
- DICOM (DCM*)
*whole-slide images

HiPlex RNAscope™ Application Note
Using our FISH module we demonstrate how to perform 12-plex RNAscope image analysis using ACD’s HiPlexv2 assay.
Submit the form below to view the requested document
RNAscope™ Image Analysis Using HALO and HALO AI
Using our ISH module for brightfield and our FISH module for fluorescence, we demonstrate how to quantify ISH signal from RNAscope™ assays based on ACD Bio’s recommended scoring guidelines.
Submit the form below to view the requested document
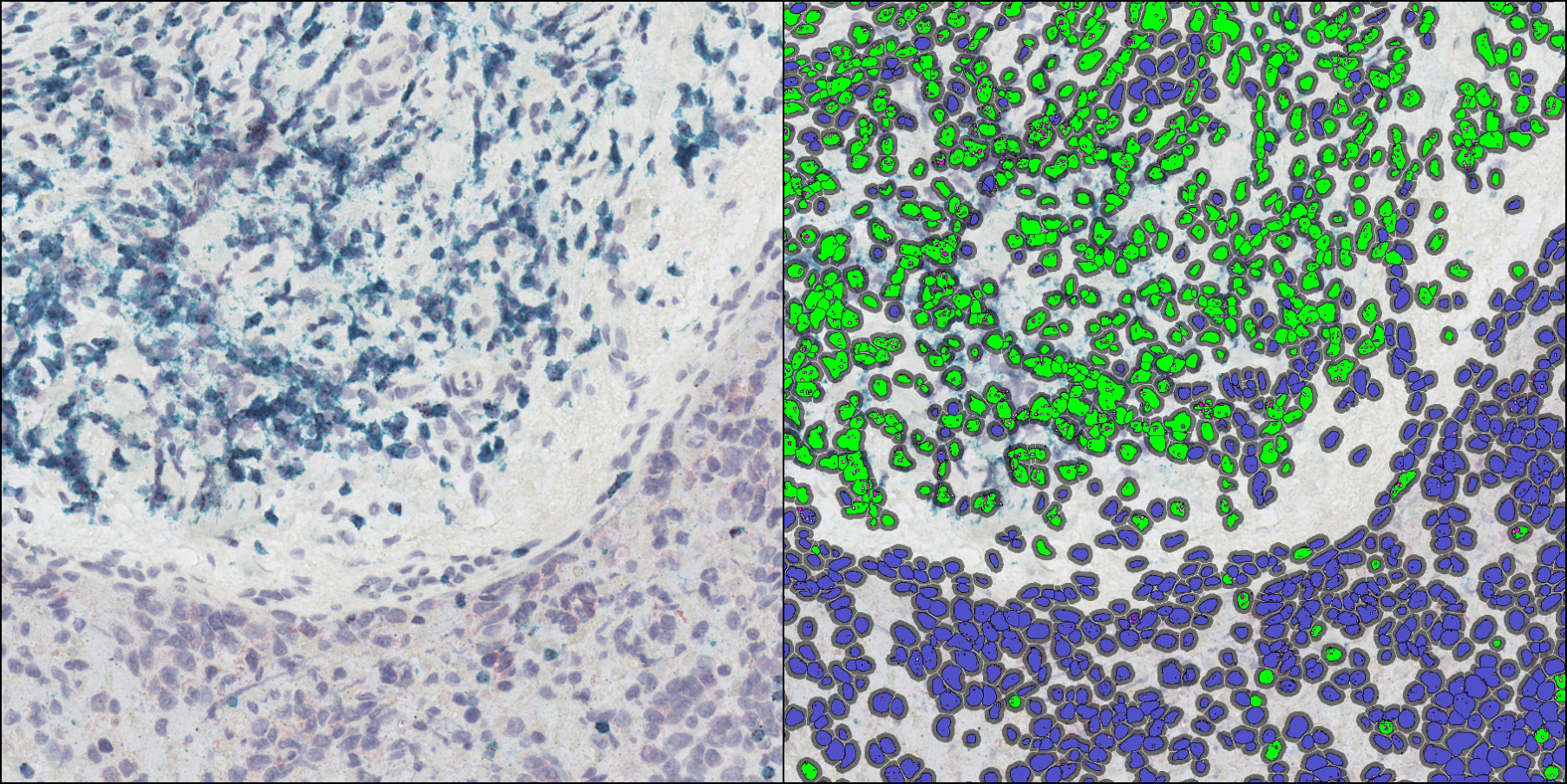
Spatial Multiplex Profiling of Immune Markers with the RNAscope Hiplex v2 Assay
Download this poster to learn about combining ACD’s HiPlex v2 in situ hybridization assay with HALO image analysis.
Submit the form below to view the requested document

Quantitative RNAscope Image Analysis Guide
From experimental design considerations to optimized setup of HALO image analysis parameters, our guide will help take your quantitative RNAscope™ image analysis to the next level.
Submit the form below to view the requested document

Hedgehog Signaling pathway in Thymic Differentiation – An in situ Single Cell Resolution Investigation
1 February 2024 | Please join us for this 1-hour webinar to Learn about applications of the ACD RNAscope™ HiPlex assay .

A role for HALO® in characterizing cell heterogeneity across organ systems: from the liver to the brain
12 October 2023 | Please join us for this 1-hour webinar to learn about characterizing cell heterogeneity with HALO to gain new biological insights.

Getting Started with RNAscope™ Image Analysis in HALO®
28 March 2023 | Join us for this 1-hour webinar to see a live demonstration of RNAscope image analysis using the HALO® platform from Indica
Masterclass Webinar: Optimizing RNAscope Image Analysis
05 May 2022 | In this 60-min webinar, Dr. Ghislaine Lioux will present solutions to common RNAscope image analysis challenges including how to optimize
Publication Spotlight
The table below includes publications that cite the FISH and ISH modules.
Your publication not on the list? Drop us an email to let us know about it!
| Title | Authors | Year | Journal | Application | HALO Modules | Product |
|---|---|---|---|---|---|---|
| EBV+†tumors exploit tumor cell-intrinsic and -extrinsic mechanisms to produce regulatory T cell-recruiting chemokines CCL17 and CCL22 | Jorapur A, Marshall L, Jacobson S, Xu M, Marubayashi S, Zibinsky M, Hu D, Robles O, Jackson J, Baloche V, Busson P, Wustrow D, Brockstedt D, Talay O, Kassner P, Cutler G | 2022 | PLOS Pathogens | Oncology | ISH/FISH | HALO |
| AAV9/MFSD8 gene therapy is effective in preclinical models of neuronal ceroid lipofuscinosis type 7 disease | Chen X, Dong T, Hu Y, Shaffo F, Belur N, Mazzulli J, Gray S | 2022 | The Journal of Clinical Investigation | Neuroscience | Area Quantification, ISH/FISH | HALO |
| Genome Sequence and Experimental Infection of Calves with Bovine Gammaherpesvirus 4 (BoHV-4) | Bauermann F, Falkenberg S, Martins M, Dassanayake R, Neill J, Ridpath J, Silveira S, Palmer M, Buysse A, Mohr A, Flores E, Diel D | 2022 | Research Square | Infectious Disease, Other | ISH/FISH | HALO |
| Vertical transmission of attaching and invasive E. coli from the dam to neonatal mice predisposes to more severe colitis following exposure to a colitic insult later in life | Brand M, Proctor A, Hostetter J, Zhou N, Friedberg I, Jergens A, Phillips G, Wannemuehler M | 2022 | PLOS ONE | Other | ISH/FISH | HALO |
| Epicardium-derived cells organize through tight junctions to replenish cardiac muscle in salamanders | Eroglu E, Yen C, Witman N, Elewa A, Araus A, Wang H, Szattler T, Umeano C, Sohlmer J, Goedel A, Simon A, Chien K | 2022 | Nature Cell Biology | Myology | ISH/FISH, ISH-IHC/FISH-IF | HALO |
| TGF?1 Induces Senescence and Attenuated VEGF Production in Retinal Pericytes | Avarmovic D, Archaimbault S, Kemble A, Gruener S, Lazendic M, Westenskow P | 2022 | Biomedicines | Other | ISH/FISH | HALO |
| Satellite repeat RNA expression in epithelial ovarian cancer associates with a tumor immunosuppressive phenotype | Porter R, Sun S, Flores M, Berzolla E, You E, Phillips I, KC N, Desai N, Tai E, Szabolcs A, Lang E, Pankaj A, Raabe M, Thapar V, Xu K, Nieman L, Rabe D, Kolin D, Stover E, Pepin D, Stott S, Deshpande V, Liu J, Solovyov A, Matulonis U, Greenbaum B, Ting D | 2022 | The Journal of Clinical Investigation | Immuno-oncology | ISH/FISH, ISH-IHC/FISH-IF | HALO |
| Distinct Encoding of Reward and Aversion by Peptidergic BNST Inputs to the VTA | Soden M, Yee J, Cuevas B, Rastani A, Elum J, Zweifel L | 2022 | Frontiers in Neural Circuits | Neuroscience | ISH/FISH | HALO |
| Hyperpolarised†13C-MRI identifies the emergence of a glycolytic cell population within intermediate-risk human prostate cancer | Sushentsev N, McLean M, Warren A, Benjamin A, Brodie C, Frary A, Gill A, Jones J, Kaggie J, Lamb B, Locke M, Miller J, Mills I, Priest A, Robb F, Shah N, Schulte R, Graves M, Gnanapragasam V, Brindle K, Barrett T, Gallacher F | 2022 | Nature Communications | Oncology | ISH/FISH, Membrane, Multiplex IHC | HALO |
| Adult re-expression of IRSp53 rescues NMDA receptor function and social behavior in IRSp53-mutant mice | Noh Y, Yook C, Kang J, Lee S, Kim Y, Yang E, Kim H, Kim E | 2022 | Communications Biology | Neuroscience | ISH/FISH, Figure Maker | HALO |
| The NALCN channel regulates metastasis and nonmalignant cell dissemination | Rahrmann E, Shorthouse D, Jassim A, Hu L, Ortiz M, Mahler-Araujo B, Vogel P, Paez-Ribes M, Fatemi A, Hannon G, Iyer R, Blundon J, Lourenco F, Kay J, Nazarian R, Hall B, Zakharenko S, Winton D, Zhu L, Gilbertson R | 2022 | Nature Genetics | Oncology | ISH/FISH, Multiplex IHC | HALO |
| Relaxin/insulin-like family peptide receptor 4 (Rxfp4) expressing hypothalamic neurons Q8 modulate food intake and preference in mice | Lewis J, Woodward O, Nuzzaci D, Smith C, Adriaenssens A, Billing L, Brighton C, Phillips B, Tadross J, Kinston S, Ciabatti E, Gottgens B, Tripodi M, Mornigold D, Baker D, Gribble F, Reimann F | 2022 | Molecular Metabolism | Metabolism | ISH/FISH, Highplex FL | HALO |
| Regional epithelial cell diversity in the small intestine of pigs | Wiarda J, Becker S, Sivasankaran S, Loving C, | 2022 | Journal of Animal Science | Gastroenterology | ISH/FISH, Figure Maker | HALO |
| Single cell analysis of cribriform prostate cancer reveals cell intrinsic and tumor microenvironmental pathways of aggressive disease | Wong H, Sheng Q, Hesterberg A, Croessmann S, Rios B, Giri K, Jackson J, Miranda A, Watkins E, Schaffer K, Donahue M, Winkler E, Penson D, Smith J, Herrell S, Luckenbaugh A, Barocas D, Kim Y, Graves D, Giannico G, Rathmell J, Park B, Gordetsky J, Hurley P | 2022 | Nature Communications | Oncology | ISH/FISH | HALO |
| Systemic Nos2 Depletion and Cox inhibition limits TNBC disease progression and alters lymphoid cell spatial orientation and density | Somasundaram V, Ridnour L, Cheng R, Walke A, Kedei N, Bhattacharyya D, Wink A, Edmondson E, Butcher D, Warner A, Dorsey T, Scheiblin D, Heinz W, Bryant R, Kinders R, Lipkowitz S, Wong S, Pore M, Hewitt S, McVicar D, Anderson S, Chang J, Glynn S, Ambs S, Lockett S, Wink D | 2022 | Redox Biology | Oncology | Classifer, ISH/FISH, Spatial Analysis, Registration, ISH-IHC/FISH-IF | HALO |
| Glutaminase 2 Knockdown Reduces Hyperammonemia and Associated Lethality of Urea Cycle Disorder Mouse Model | Mao X, Chen H, Lin A, Kim S, Burczynski M, Na E, Halasz G, Sleeman M, Murphy A, Okamoto H, Cheng X | 2022 | Journal of Inherited Metabolic Disease | Metabolism | Area Quantification, ISH/FISH | HALO |
| What Happens to the Preserved Renal Parenchyma After Clamped Partial Nephrectomy? | Xiong L, Nguyen J, Peng Y, Zhou Z, Ning K, Jia N, Nie J, Wen D, Wu Z, Roversi G, Palacios D, Abramczyk E, Munoz-Lopez C, Campbell J, Cao Y, Li Wencai, Zhang X, He Z, Li X, Huang J, Shou J, Wu J, Chen M, Chen X, Zheng J, Xu C, Zhong W, Li Z, Dong W, Zhou J, Zhang H, Luo J, Liu J, Sun G, Han H, Guo S, Dong P, Zhou F, Yu C, Campbell S, Zhang Z | 2022 | European Urology | Other | Area Quantification, ISH/FISH | HALO |
| Adolescent intermittent ethanol exposure produces Sex-Specific changes in BBB Permeability: A potential role for VEGFA | Vore A, Barney T, Deak M, Varlinskaya E, Deak T | 2022 | Brain, Behavior, and Immunity | Neuroscience | ISH/FISH | HALO |
| Relationship between impaired BMP signalling and clinical risk factors at early-stage vascular injury in the preterm infant | Heydarian M, Oak P, Zhang X, Kamgari N, Kindt A, Koschlig M, Pritzke T, Gonzalez-Rodriguez E, Forster K, Morty R, Hafner F, Hubener C, Flemmer A, Yildirim A, Sudheendra D, Tian X, Petrera A, Kirsten H, Ahnert P, Morrell N, Desai T, Sucre J, Spiekerkoetter E, Hilgendorff A | 2022 | Thorax | Other | ISH/FISH | HALO |
| Spatiotemporal expression pattern of Progesterone Receptor Component (PGRMC) 1 in endometrium from patients with or without endometriosis or adenomyosis | Thieffry C, Van Wynendaele M, Samain L, Tyteca D, Pierreux C, Marbaix E, Henriet P | 2022 | The Journal of Steroid Biochemistry and Molecular Biology | Other | ISH/FISH, Highplex FL | HALO, HALO AI |
| Quantitative Imaging Analysis of the Spatial Relationship between Antiretrovirals, Reverse Transcriptase Simian-Human Immunodeficiency Virus RNA, and Fibrosis in the Spleens of Nonhuman Primates | Devanathan A, White N, Desyaterik Y, De la Cruz G, Nekorchuk M, Terry M, Busman-Sahay K, Adamson L, Luciw P, Fedoriw Y, Estes J, Rosen E, Kashuba A | 2022 | Antimicrobial Agents and Chemotherapy | Infectious Disease | ISH/FISH | HALO |
| CD123-directed allogeneic chimeric-antigen receptor T-cell therapy (CAR-T) in blastic plasmacytoid dendritic cell neoplasm (BPDCN): Clinicopathological insights | Pemmaraju N, Wilson N, Senapati J, Economides M, Guzman M, Neelapu S, Kazemimood R, Davis R, Jain N, Khoury J, Sugita M, Cai T, Smith J, Frattini M, Garton A, Roboz G, Konopleva M | 2022 | Leukemia Research | Immuno-oncology | ISH/FISH | HALO |
| Localization of macrophage subtypes and neutrophils in the prostate tumor microenvironment and their association with prostate cancer racial disparities | Maynard J, Godwin T, Lu J, Vidal I, Lotan T, De Marzo A, Joshu C, Sfanos K | 2022 | The Prostate | Oncology | ISH/FISH, Multiplex IHC | HALO |
| Molecular identification of spatially distinct anabolic responses to mechanical loading in murine cortical bone | Chlebek C, Moore J, Ross F, van der Meulen M | 2022 | Journal of Bone and Mineral Research | Other | ISH/FISH | HALO |
| Sex-specific effects of ethanol consumption in older Fischer 344 rats on microglial dynamics and A_(1-42) accumulation | Marsland P, Vore A, DaPrano E, Paluch J, Blackwell A, Varlinskaya E, Deak T | 2022 | Alcohol | Neuroscience | ISH/FISH | HALO |
| Estrogen receptor beta activity contributes to both tumor necrosis factor alpha expression in the hypothalamic paraventricular nucleus and the resistance to hypertension following angiotensin II in female mice | Woods C, Contoreggi N, Johnson M, Milner T, Wang G, Glass M, | 2022 | Neurochemistry International | Neuroscience | ISH/FISH | HALO |
| Angiotensin II Infusion Results in Both Hypertension and Increased AMPA GluA1 Signaling in Hypothalamic Paraventricular Nucleus of Male but not Female Mice | Wang G, Woods C, Johnson M, Milner T, Glass M | 2022 | Neuroscience | Neuroscience | ISH/FISH | HALO |
| Multiregional single-cell dissection of tumor and immune cells reveals stable lock-and-key features in liver cancer | Ma L, Heinrich S, Wang L, Keggenhoff F, Khatib S, Forgues M, Kelly M, Hewitt S, Saif A, Hernandez J, Mabry D, Kloeckner R, Greten T, Chaisaingmongkol J, Ruchirawat M, Marquardt J, Wang X | 2022 | Nature Communications | Oncology | ISH/FISH | HALO |
| Autologous T cell therapy for MAGE-A4+ solid cancers in HLA-A*02+ patients: a phase 1 trial | Hong D, Van Tine B, Biswas S, McAlpine C, Johnson M, Olszanski A, Clarke J, Araujo D, Blumenschein G, Kebriaei P, Lin Q, Tipping A, Sanderson J, Wang R, Trivedi T, Annareddy T, Bai J, Rafail S, Sun A, Fernandes L, Navenot J-M, Bushman F, Everett J, Karadeniz D, Broad R, Isabelle M, Naidoo R, Bath N, Betts G, Wolchinsky Z, Batrakou D, Van Winkle E, Elefant E, Ghobadi A, Cashen A, Grand'Maison A, McCarthy P, Fracasso P, Norry E, Williams D, Druta M, Liebner D, Odunsi K, Butler M | 2023 | Nature Medicine | Immuno-oncology | ISH/FISH, Spatial Analysis, Highplex FL | HALO |
| Combined KRASG12C and SOS1 inhibition enhances and extends the anti-tumor response in KRASG12C-driven cancers by addressing intrinsic and acquired resistance | Thatikonda V, Lu H, Jurado S, Kostyrko K, Bristow C, Bosch K, Feng N, Gao S, Gerlach D, Gmachi M, Lieb S, Jeschko A, Machado A, Marszalek E, Mahendra M, Jaeger P, Sorokin A, Strauss S, Trapani F, Kopetz S, Vellano C, Petronczki M, Kraut N, Heffernan T, Marszalek J, Pearson M, Waizenegger I, Hofmann M | 2023 | bioRxiv | Oncology | ISH/FISH, Multiplex IHC, Highplex FL, Figure Maker | HALO |
| Prefrontal cortical protease TACE/ADAM17 is involved in neuroinflammation and stress-related eating alterations | Sharafeddin F, Ghaly M, Simon T, Ontiveros-Angel P, Figueroa J | 2023 | bioRxiv | Neuroscience | ISH/FISH | HALO |
| Long noncoding RNA LINC01594 inhibits the CELF6-mediated splicing of oncogenic CD44 variants to promote colorectal cancer metastasis. | Liu B, Song A, Gui P, Wang J, Pan Y, Li C, Li S, Zhang Y, Jiang T, Xu Y, Huo F, Pei D, Song J | 2023 | Research Square | Oncology | ISH/FISH | HALO |
| CD47-Dependent Regulation of Immune Checkpoint Gene Expression and MYCN mRNA Splicing in Murine CD8 and Jurkat T Cells | Kaur S, Awad D, Finney R, Meyer T, Singh S, Cam M, Karim B, Warner A, Roberts D | 2023 | International Journal of Molecular Sciences | Immuno-oncology | ISH/FISH | HALO |
| SARS-CoV-2 infection induces DNA damage, through CHK1 degradation and impaired 53BP1 recruitment, and cellular senescence | Gioia U, Tavella S, Martinez-Orellana P, Cicio G, Colliva A, Ceccon M, Cabrini M, Henriques A, Fumagalli V, Paldino A, Presot E, Rajasekharan S, Iacomino N, Pisati F, Matti V, Sepe S, Conte M, Barozzi S, Lavagnino Z, Carletti T, Volpe M, Cavalcante P, Iannacone M, Rampazzo C, Bussani R, Tripodo C, Zacchigna S, Marcello A, di Fagagna F | 2023 | Nature Cell Biology | Infectious Disease | ISH/FISH, Highplex FL | HALO |
| A cellular and molecular spatial atlas of dystrophic muscle | Stec M, Su Q, Adler C, Zhang L, Golann D, Khan N, Panagis L, Villalta S, Ni M, Wei Y, Walls J, Murphy A, Yancopoulos G, Atwal G, Kleiner S, Halasz G, Sleeman M | 2023 | Proceedings of the National Academy of Sciences | Myology | ISH/FISH, Highplex FL | HALO |
Related HALO Modules
Separate multiple tissue classes across a tissue using a learn-by-example approach. Can be used in conjunction with all other modules (fluorescent and brightfield) to select specific tissue classes for further analysis.
Learn MoreQuantify spot number, optical density and co-expression of one or two brightfield RNA or DNA probes on a per cell basis. Includes support for brightfield RNAscope assays.
Learn MoreQuantify IHC positivity and spot number, optical density and co-expression of one or two brightfield RNA or DNA probes on a per cell basis. Includes support for brightfield RNAscope assays.
Learn MoreUse the arrows above to view additional related modules
Want to Learn More?
Fill out the form below to request information about any of our software products.
You can also drop us an email at info@indicalab.com