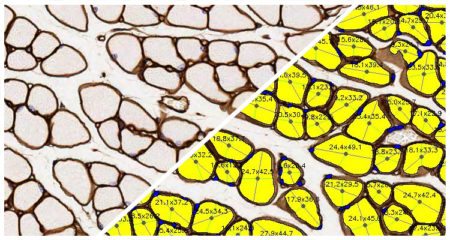
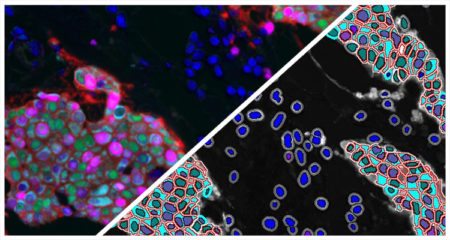
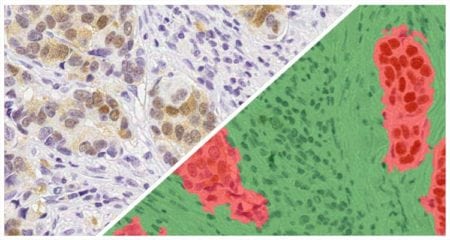
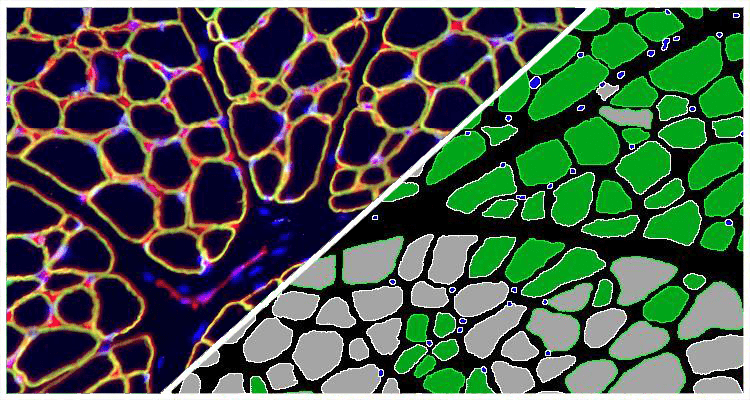
The Muscle Fiber module detects fibers and reports the total count of muscle fibers as well as the perimeter, area, and diameter of each fiber.
Since muscle fiber cross-sections are irregularly shaped, the algorithm uses patent pending image processing techniques to detect each fiber’s minimum diameter (green lines) and maximum diameter (blue lines). Finally the software reports the diameter (average/minimum/maximum), the average perimeter length, and the average fiber area for all fibers in the entire section.
File formats supported by the HALO image analysis platform:
- Non-proprietary (JPG, TIF, OME.TIFF)
- Nikon (ND2)
- 3D Histech (MRXS)
- Akoya (QPTIFF, component TIFF)
- Olympus / Evident (VSI)
- Hamamatsu (NDPI, NDPIS)
- Aperio (SVS, AFI)
- Zeiss (CZI)
- Leica (SCN, LIF)
- Ventana (BIF)
- Philips (iSyntax, i2Syntax)
- KFBIO (KFB, KFBF)
- DICOM (DCM*)
*whole-slide images

HALO® MASTERCLASS WEBINAR SERIES
Fall/Winter of 2020 | Indica Labs is excited to continue our HALO® Masterclass Webinar Series this autumn. Each masterclass webinar will offer a deep dive
Publication Spotlight
The table below includes publications that cite the Muscle Fiber and Muscle Fiber FL modules.
Your publication not on the list? Drop us an email to let us know about it!
| Title | Authors | Year | Journal | Application | HALO Modules | Product |
|---|---|---|---|---|---|---|
| Inhibition of myostatin prevents microgravity-induced loss of skeletal muscle mass and strength | Smith RC, Cramer MS, Mitchell PJ, Lucchesi J, Ortega AM, Livingston EW, Ballard D, Zhang L, Hanson J, Barton K, Berens S, Credille KM, Bateman TA, Ferguson VL, Ma YL, Stodieck LS | 2020 | PLOS One | Myology | Muscle Fiber | HALO |
| Dynamic changes to lipid mediators support transitions among macrophage subtypes during muscle regeneration | Giannakis N, Sansbury BE, Patsalos A, Hays TT, Riley CO, Han X, Spite M, Nagy L | 2019 | Nature Immunology | Immunology, Myology | Muscle Fiber | HALO |
| Myostatin blockade with a fully human monoclonal antibody induces muscle hypertrophy and reverses muscle atrophy in young and aged mice | Latres E, Pangilinan J, Miloscio L, Bauerlein R, Na E, Potocky TB, Huang Y, Eckersdorff M, Rafique A, Mastaitis J, Lin C, Murphy AJ, Yancopoulos GD, Gromada J, Stitt T | 2015 | Skeletal Muscle | Myology | Muscle Fiber | HALO |
| Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy | Lieberman AP, Yu Z, Murray S, Peralta R, Low A, Guo S, Yu XX, Cortes CJ, Bennett CF, Monia BP, La Spada AR, Hung G | 2014 | Cell Reports | Myology | Muscle Fiber | HALO |
| The Goto Kakizaki rat: Impact of age upon changes in cardiac and renal structure, function | Meagher P, Civitarese R, Lee X, Gordon M, Bugyei-Twum A, Desjardins J, Kabir G, Zhang Y, Kosanam H, Visram A, Leong-Poi H, Advani A, Connelly K | 2022 | PLOS ONE | Myology | Muscle Fiber | HALO |
| A growth factorñexpressing macrophage subpopulation orchestrates regenerative inflammation via GDF-15 | Patsalos A, Halasz L, Medina-Serpas M, Berger W, Daniel B, Tzerpos P, Kiss M, Nagy G, Fischer C, Simandi Z, Varga T, Nagy L | 2021 | Journal of Experimental Medicine | Muscle Fiber | HALO | |
| Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice | Kenjo E, Hozumi H, Makita Y, Iwabuchi K, Fujimoto N, Matsumoto S, Kimura M, Amano Y, Ifuku M, Naoe Y, Inukai N, Hotta A | 2021 | Nature Communications | Other | Muscle Fiber | HALO |
| Electrical impedance myography detects dystrophin-related muscle changes in mdx mice | Hiyoshi T, Zhao F, Baba R, Hirakawa T, Kuboki R, Suzuki K, Tomimatsu Y, O'Donnell P, Han S, Zach N, Nakashima M | 2023 | Research Square | Myology | Area Quantification, Muscle Fiber | HALO |
| Effects of the purified dry extract of fermented ginseng BST204 on muscle fiber regeneration | Jo S, Park Y, Chang Y, Moon J, Lee S, Lee H, Kim M, Kim D, Bae S, Park S, Yun H, You J, Im M, Han H, Kim S, Jin D | 2023 | Biochemistry and Biophysics Reports | Myology | Muscle Fiber | HALO |
| Rotator cuff muscle fibrosis can be assessed using ultrashort echo time magnetization transfer MRI with fat suppression | Chang EY, Suprana A, Tang Q, Cheng X, Fu E, Orozco E, Jerban S, Shah S, Du J, Ma Y | 2023 | NMR in Biomedicine | Fibrosis | Muscle Fiber | HALO |
| Rotator cuff muscle fibrosis can be assessed using ultrashort echo time magnetization transfer MRI with fat suppression | Chang EY, Suprana A, Tang Q, Cheng X, Fu E, Orozco E, Jerban S, Shah S, Du J, Ma Y | 2023 | NMR in Biomedicine | Fibrosis | Muscle Fiber | HALO |
| Peripheral neural interfaces: Skeletal muscles are hyper-reinnervated according to the axonal capacity of the surgically rewired nerves | Tereshenko V, Dotzauer D C, Schmoll M, Harnoncourt L, Carrero Rojas G, Gfrerer L, Eberlin K R, Austen Jr. W G, Blumer R, Farina D, Aszmann O C | 2024 | Science Advances | Neuroscience | Muscle Fiber | HALO |
Related HALO Module
Quantify expression of an unlimited number of biomarkers in any cellular compartment - membrane, nucleus or cytoplasm.
Learn MoreSeparate multiple tissue classes across a tissue using a learn-by-example approach. Can be used in conjunction with all other modules (fluorescent and brightfield) to select specific tissue classes for further analysis.
Learn MoreQuantify positive fiber or membrane staining for an unlimited number of channels.
Learn MoreUse the arrows above to view additional related modules
Want to Learn More?
Fill out the form below to request information about any of our software products.
You can also drop us an email at info@indicalab.com